Workshop ChIPATAC 2020
Computational analysis of ChIP-seq and ATAC-seq data
14-15 December 2020ChIP-seq and ATAC-seq Bioinformatics Analysis Workshop
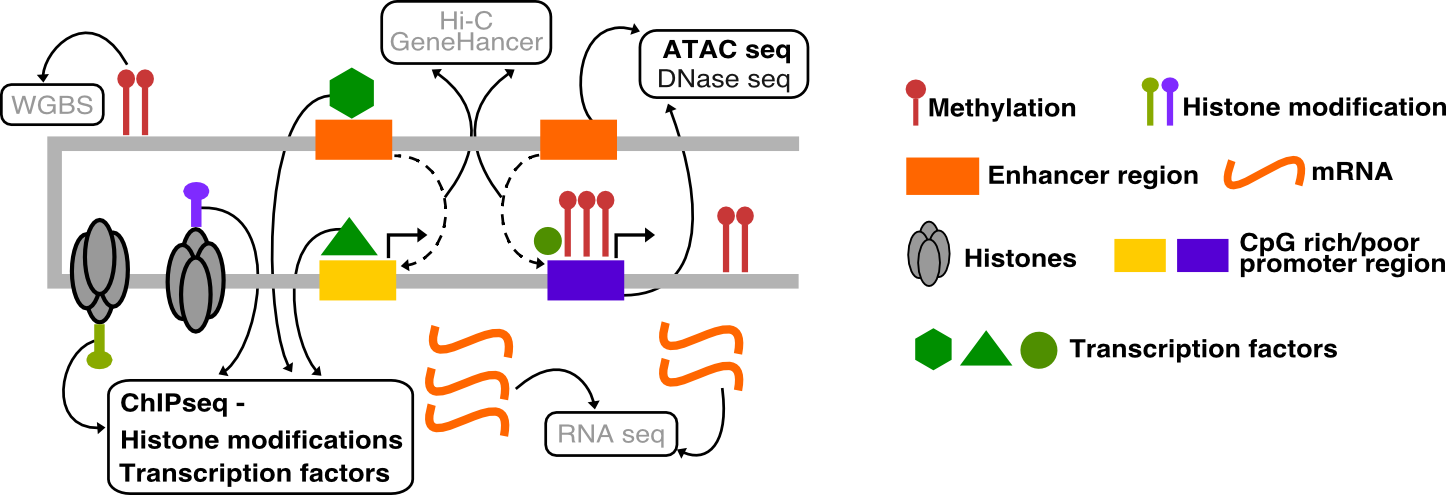
Welcome to the workshop on the computational analysis of ChIP-seq and ATAC-seq data. This workshop is meant for individuals with little to no previous knowledge of bioinformatics. We will guide you through each of the main steps of the bioinformatic analysis.
Tutors
- Carl Herrmann, Health Data Science Unit Medical Faculty Heidelberg and BioQuant (carl.herrmann@bioquant.uni-heidelberg.de)
- Ashwini Sharma, Health Data Science Unit Medical Faculty Heidelberg and BioQuant (ashwini.sharma@bioquant.uni-heidelberg.de)
Slides
- Part 1 - general intro
- Part 2 - ChIP : QC, trimming, alignments
- Part 3 - ChIP : peak calling, annotation, signal tracks, QC
- Part 4 - ChIP : ATAC-seq
Practical parts
- General instructions before you start
- Make sure to protocol your progress in this Google Sheet
Day 1: ChIP-seq
- Introduction
- Quality control on reads
- Read trimming
- Read alignment
- Peak calling
- Peak annotation
- ChIP-seq quality control
- Generating signal tracks and using IGV
Day 2: ATAC-seq
Due to time constraints, please start with part 5, and skip parts 1-4 for the moment!!
Day 2: Going further: integrating/comparing ChIP-seq/ATAC-seq
Schedule
The course will run over 2 days (Monday, 14.12 and Tuesday, 15.12) from 10 am - 12.30 pm and 1.30 pm - 5.30 pm.
Check the detailed schedule here
Organisation
The course will be online only!
- Lectures will be over Zoom (use this link)
- We will use Discord channels for the practical sessions (register using this link)
Technical pre-requisites
All the computation will be done on the cloud of the German Bioinformatics Network de.NBI. All you need is a personal laptop to access the cloud server.
Check the technical pre-requisites, depending on whether your operating system is Windows, MacOS or Linux